Contents
gmixcv.m
From A First Course in Machine Learning, Chapter 6. Simon Rogers, 01/11/11 [simon.rogers@glasgow.ac.uk] Fitting a Gaussian mixture
clear all;close all; path(path,'../utilities');
Load the data
load ../data/kmeansdata
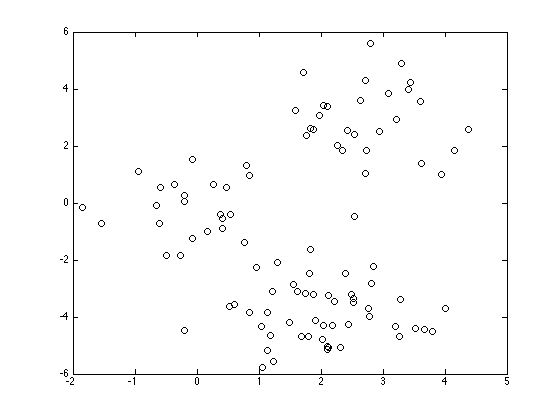
Plot the data
figure(1);hold off plot(X(:,1),X(:,2),'ko');
Do a 10-fold CV
NFold = 10;
N = size(X,1);
sizes = repmat(floor(N/NFold),1,NFold);
sizes(end) = sizes(end) + N - sum(sizes);
csizes = [0 cumsum(sizes)];
order = randperm(N); % Randomise the data order
MaxIts = 100;
Loop over K
Kvals = 1:10; for kv = 1:length(Kvals) K = Kvals(kv); for fold = 1:NFold fprintf('\nK: %g, fold: %g',K,fold); foldindex = order(csizes(fold)+1:csizes(fold+1)); trainX = X; trainX(foldindex,:) = []; testX = X(foldindex,:); % Train the mixture means = randn(K,2); for k = 1:K covs(:,:,k) = rand*eye(2); end priors = repmat(1/K,1,K); B = []; B(1) = -inf; converged = 0; it = 0; tol = 1e-2; Ntrain = size(trainX,1); D = size(X,2); while 1 it = it + 1; % Update q temp = zeros(Ntrain,K); for k = 1:K const = -(D/2)*log(2*pi) - 0.5*log(det(covs(:,:,k))); Xm = trainX - repmat(means(k,:),Ntrain,1); temp(:,k) = const - 0.5 * diag(Xm*inv(covs(:,:,k))*Xm'); end % Compute the Bound on the likelihood if it>1 B(it) = sum(sum(q.*log(repmat(priors,Ntrain,1)))) + ... sum(sum(q.*temp)) - ... sum(sum(q.*log(q))); if abs(B(it)-B(it-1))<tol converged = 1; end end if converged == 1 || it>MaxIts break end temp = temp + repmat(priors,Ntrain,1); q = exp(temp - repmat(max(temp,[],2),1,K)); % Minor hack for numerical issues - stops the code crashing when % clusters are empty q(q<1e-3) = 1e-3; q(q>1-1e-3) = 1-1e-3; q = q./repmat(sum(q,2),1,K); % Update priors priors = mean(q,1); % Update means for k = 1:K means(k,:) = sum(trainX.*repmat(q(:,k),1,D),1)./sum(q(:,k)); end % update covariances for k = 1:K Xm = trainX - repmat(means(k,:),Ntrain,1); covs(:,:,k) = (Xm.*repmat(q(:,k),1,D))'*Xm; covs(:,:,k) = covs(:,:,k)./sum(q(:,k)); end end % Compute the held-out likelihood Ntest = size(testX,1); temp = zeros(Ntest,K); for k = 1:K const = -(D/2)*log(2*pi) - 0.5*log(det(covs(:,:,k))); Xm = testX - repmat(means(k,:),Ntest,1); temp(:,k) = const - 0.5 * diag(Xm*inv(covs(:,:,k))*Xm'); end temp = exp(temp).*repmat(priors,Ntest,1); outlike(kv,csizes(fold)+1:csizes(fold+1)) = log(sum(temp,2))'; end end
K: 1, fold: 1 K: 1, fold: 2 K: 1, fold: 3 K: 1, fold: 4 K: 1, fold: 5 K: 1, fold: 6 K: 1, fold: 7 K: 1, fold: 8 K: 1, fold: 9 K: 1, fold: 10 K: 2, fold: 1 K: 2, fold: 2 K: 2, fold: 3 K: 2, fold: 4 K: 2, fold: 5 K: 2, fold: 6 K: 2, fold: 7 K: 2, fold: 8 K: 2, fold: 9 K: 2, fold: 10 K: 3, fold: 1 K: 3, fold: 2 K: 3, fold: 3 K: 3, fold: 4 K: 3, fold: 5 K: 3, fold: 6 K: 3, fold: 7 K: 3, fold: 8 K: 3, fold: 9 K: 3, fold: 10 K: 4, fold: 1 K: 4, fold: 2 K: 4, fold: 3 K: 4, fold: 4 K: 4, fold: 5 K: 4, fold: 6 K: 4, fold: 7 K: 4, fold: 8 K: 4, fold: 9 K: 4, fold: 10 K: 5, fold: 1 K: 5, fold: 2 K: 5, fold: 3 K: 5, fold: 4 K: 5, fold: 5 K: 5, fold: 6 K: 5, fold: 7 K: 5, fold: 8 K: 5, fold: 9 K: 5, fold: 10 K: 6, fold: 1 K: 6, fold: 2 K: 6, fold: 3 K: 6, fold: 4 K: 6, fold: 5 K: 6, fold: 6 K: 6, fold: 7 K: 6, fold: 8 K: 6, fold: 9 K: 6, fold: 10 K: 7, fold: 1 K: 7, fold: 2 K: 7, fold: 3 K: 7, fold: 4 K: 7, fold: 5 K: 7, fold: 6 K: 7, fold: 7 K: 7, fold: 8 K: 7, fold: 9 K: 7, fold: 10 K: 8, fold: 1 K: 8, fold: 2 K: 8, fold: 3 K: 8, fold: 4 K: 8, fold: 5 K: 8, fold: 6 K: 8, fold: 7 K: 8, fold: 8 K: 8, fold: 9 K: 8, fold: 10 K: 9, fold: 1 K: 9, fold: 2 K: 9, fold: 3 K: 9, fold: 4 K: 9, fold: 5 K: 9, fold: 6 K: 9, fold: 7 K: 9, fold: 8 K: 9, fold: 9 K: 9, fold: 10 K: 10, fold: 1 K: 10, fold: 2 K: 10, fold: 3 K: 10, fold: 4 K: 10, fold: 5 K: 10, fold: 6 K: 10, fold: 7 K: 10, fold: 8 K: 10, fold: 9 K: 10, fold: 10
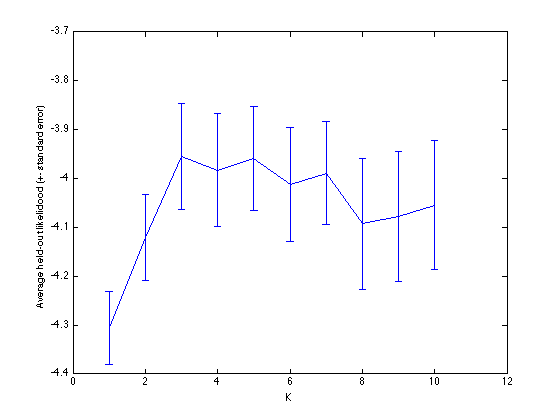
Plot the evolution in log likelihood
errorbar(Kvals,mean(outlike,2),std(outlike,1,2)./sqrt(N)); xlabel('K'); ylabel('Average held-out likelidood (+- standard error)');