Contents
cv_demo.m
From A First Course in Machine Learning, Chapter 1. Simon Rogers, 31/10/11 [simon.rogers@glasgow.ac.uk] Demonstration of cross-validation for model selection
clear all;close all;
Generate some data
Generate x between -5 and 5
N = 100;
x = 10*rand(N,1) - 5;
t = 5*x.^3 - x.^2 + x + 150*randn(size(x));
testx = [-5:0.01:5]'; % Large, independent test set
testt = 5*testx.^3 - testx.^2 + testx + 150*randn(size(testx));
Run a cross-validation over model orders
maxorder = 7; X = []; testX = []; K = 10 %K-fold CV sizes = repmat(floor(N/K),1,K); sizes(end) = sizes(end) + N - sum(sizes); csizes = [0 cumsum(sizes)]; % Note that it is often sensible to permute the data objects before % performing CV. It is not necessary here as x was created randomly. If % it were necessary, the following code would work: % order = randperm(N); % x = x(order); Or: X = X(order,:) if it is multi-dimensional. % t = t(order); for k = 0:maxorder X = [X x.^k]; testX = [testX testx.^k]; for fold = 1:K % Partition the data % foldX contains the data for just one fold % trainX contains all other data foldX = X(csizes(fold)+1:csizes(fold+1),:); trainX = X; trainX(csizes(fold)+1:csizes(fold+1),:) = []; foldt = t(csizes(fold)+1:csizes(fold+1)); traint = t; traint(csizes(fold)+1:csizes(fold+1)) = []; w = inv(trainX'*trainX)*trainX'*traint; fold_pred = foldX*w; cv_loss(fold,k+1) = mean((fold_pred-foldt).^2); ind_pred = testX*w; ind_loss(fold,k+1) = mean((ind_pred - testt).^2); train_pred = trainX*w; train_loss(fold,k+1) = mean((train_pred - traint).^2); end end
K =
10
Plot the results
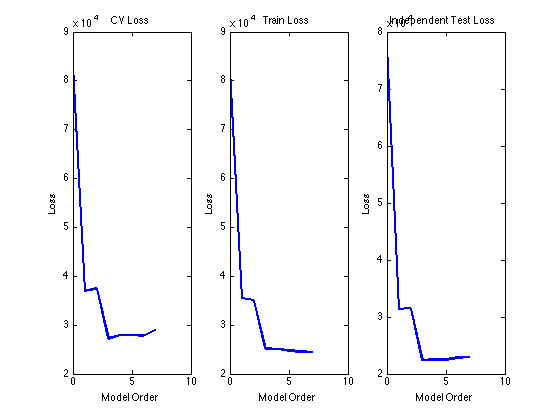
figure(1); subplot(131) plot(0:maxorder,mean(cv_loss,1),'linewidth',2) xlabel('Model Order'); ylabel('Loss'); title('CV Loss'); subplot(132) plot(0:maxorder,mean(train_loss,1),'linewidth',2) xlabel('Model Order'); ylabel('Loss'); title('Train Loss'); subplot(133) plot(0:maxorder,mean(ind_loss,1),'linewidth',2) xlabel('Model Order'); ylabel('Loss'); title('Independent Test Loss')