Contents
badmh.m
Script demonstrating issues related to MH sampling when there is high correlation
From A First Course in Machine Learning Simon Rogers, August 2016 [simon.rogers@glasgow.ac.uk]
clear all; close all;
Create the Gaussian and contours for plotting
2d Gaussian with hich correlation
mu = [1;2]; co = [1 0.95;0.95 1]; [Xv,Yv] = meshgrid([-2:0.01:4],[-1:0.01:5]); Ci = inv(co); P = zeros(size(Xv)); for i = 1:size(Xv(:)) P(i) = -log(2*pi)- 0.5*log(det(co)) - 0.5*([Xv(i)-mu(1) Yv(i)-mu(2)]*Ci*[Xv(i)-mu(1) Yv(i)-mu(2)]'); end P = exp(P); contour(Xv,Yv,P,'k','linewidth',2,'color',[0.6 0.6 0.6])
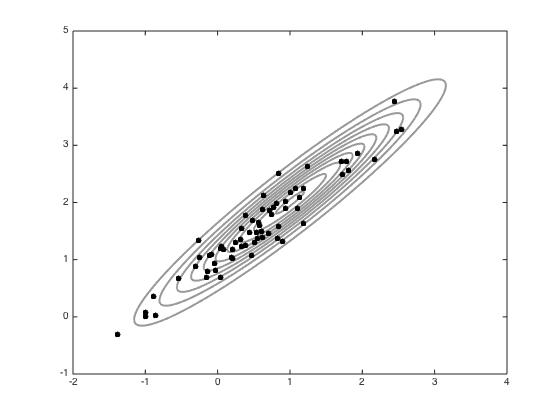
Standard MH sampling
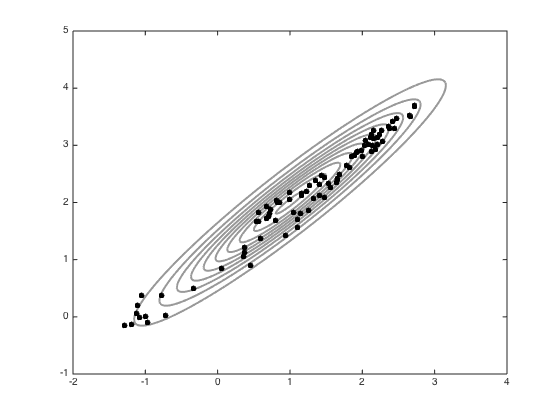
% Define the jump variance and the number of samples jss = 0.1; S = 100; all_samps = zeros(S+1,2); % Starting point x = [-1;0]; accept = 0; coI = inv(co); all_samps(1,:) = x'; % Do the sampling for s = 1:S new_x = x + randn(2,1).*sqrt(jss); new_like = -0.5*(new_x-mu)'*coI*(new_x-mu); old_like = -0.5*(x-mu)'*coI*(x-mu); r = exp(new_like - old_like); if rand<r x = new_x; accept = accept+1; end all_samps(s+1,:) = x'; end % plot the samples n_plot = 100; figure(1);hold off contour(Xv,Yv,P,'k','linewidth',2,'color',[0.6 0.6 0.6]) hold on plot(all_samps(1,1),all_samps(1,2),'ko','markersize',5,'markerfacecolor','k') for i = 2:n_plot plot(all_samps(i,1),all_samps(i,2),'ko','markersize',5,'markerfacecolor','k') end
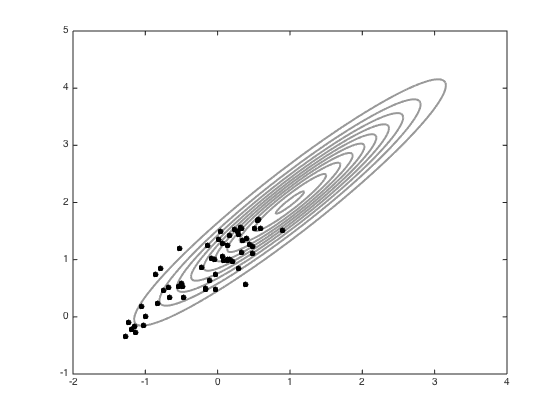
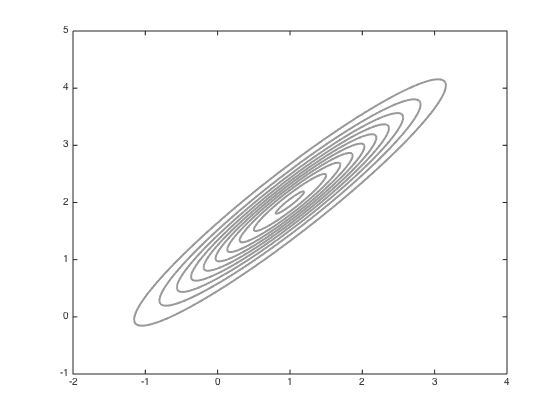
Now try adapting the proposal
Compute the sample covariance
Cs = 0.2*cov(all_samps); figure();hold off % Contour the gaussian and the new jump covariance contour(Xv,Yv,P,'k','linewidth',2,'color',[0.6 0.6 0.6]) Csi = inv(Cs); % Arbitrary mean point for the jump cov muj = [0.5;1]; Ps = zeros(size(Xv)); for i = 1:size(Xv(:)) Ps(i) = -log(2*pi)- 0.5*log(det(Cs)) - 0.5*([Xv(i)-muj(1) Yv(i)-muj(2)]*Csi*[Xv(i)-muj(1) Yv(i)-muj(2)]'); end Ps = exp(Ps); hold on contour(Xv,Yv,Ps,'k','linewidth',2) % Sample with this new covariance S = 100; all_samps_S = zeros(S+1,2); x = [-1;0]; accept_S = 0; coI = inv(co); all_samps_S(1,:) = x'; for s = 1:S new_x = x + mvnrnd(repmat(0,2,1),Cs,1)'; new_like = -0.5*(new_x-mu)'*coI*(new_x-mu); old_like = -0.5*(x-mu)'*coI*(x-mu); r = exp(new_like - old_like); if rand<r x = new_x; accept_S = accept_S + 1; end all_samps_S(s+1,:) = x'; end % plot the samples n_plot = 100; figure();hold off contour(Xv,Yv,P,'k','linewidth',2,'color',[0.6 0.6 0.6]) hold on plot(all_samps_S(1,1),all_samps_S(1,2),'ko','markersize',5,'markerfacecolor','k') for i = 2:n_plot plot(all_samps_S(i,1),all_samps_S(i,2),'ko','markersize',5,'markerfacecolor','k') end
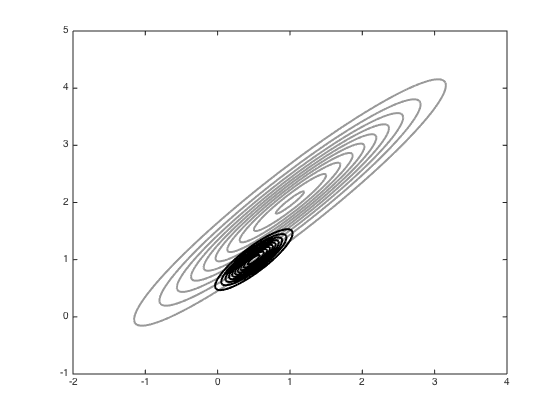
Next try Hamiltonian MCMC
Initialise parameters
M = eye(2); iM = inv(M); epsilon = 0.1; L = 10; x = [-1;0]; phi = mvnrnd(repmat(0,2,1),M,1)'; all_samps_H = zeros(S+1,2); all_samps_H(1,:) = x'; accept_H = 0; for s = 1:S new_phi = mvnrnd(repmat(0,2,1),M,1)'; new_x = x; new_phi = new_phi + 0.5*epsilon*coI*(mu-new_x); for l = 1:L-1 new_x = new_x + epsilon*iM*new_phi; new_phi = new_phi + epsilon*coI*(mu-new_x); end new_phi = new_phi + 0.5*epsilon*coI*(mu-new_x); new_like = -0.5*(new_x-mu)'*coI*(new_x-mu); old_like = -0.5*(x-mu)'*coI*(x-mu); new_like = new_like - 0.5*new_phi'*iM*new_phi; old_like = old_like - 0.5*phi'*iM*phi; r = exp(new_like - old_like); if rand<r x = new_x; accept_H = accept_H + 1; end all_samps_H(s+1,:) = x'; end % plot the samples from HMC n_plot = 100; figure();hold off contour(Xv,Yv,P,'k','linewidth',2,'color',[0.6 0.6 0.6]) hold on plot(all_samps_H(1,1),all_samps_H(1,2),'ko','markersize',5,'markerfacecolor','k') for i = 2:n_plot plot(all_samps_H(i,1),all_samps_H(i,2),'ko','markersize',5,'markerfacecolor','k') end